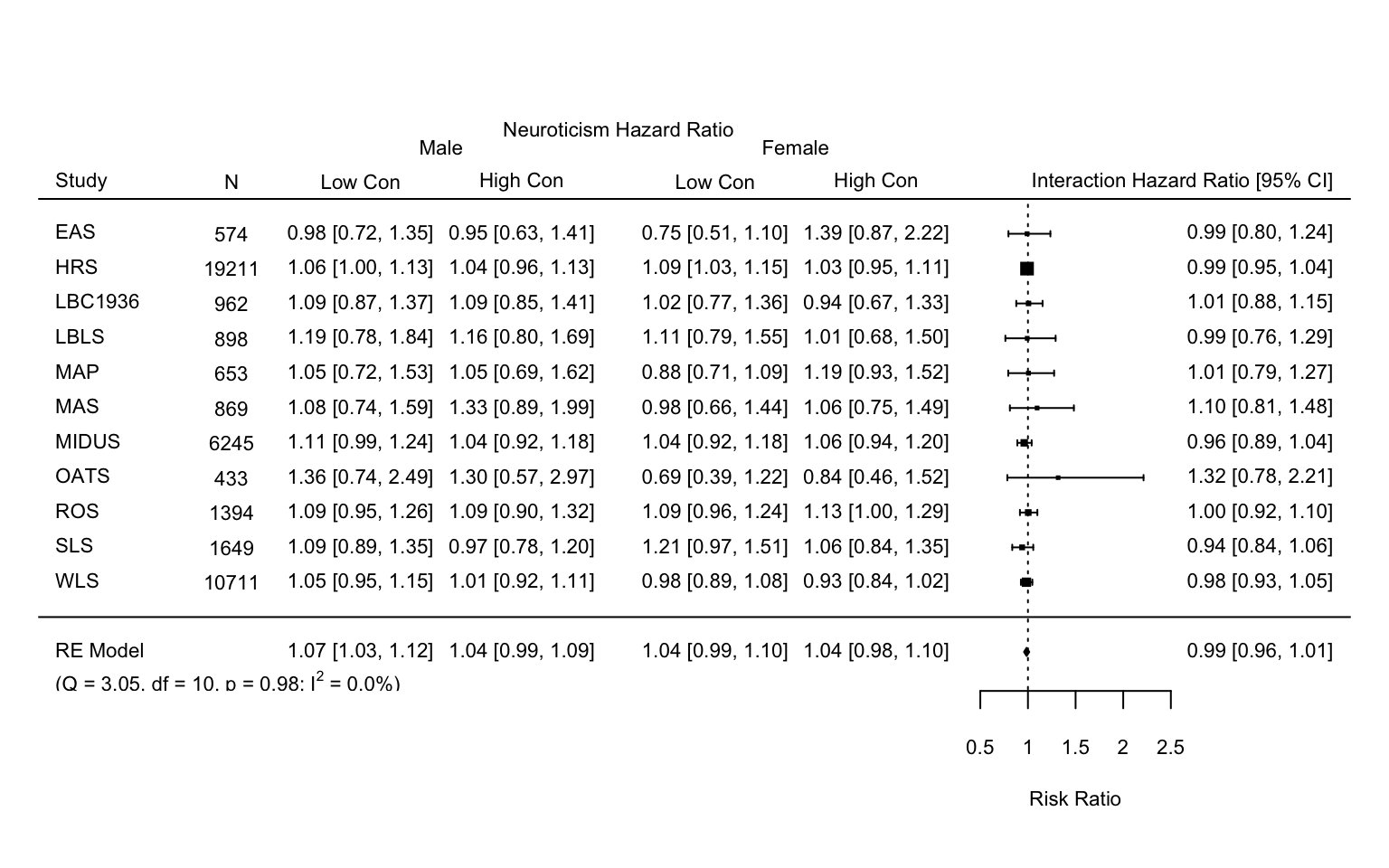
Meta analysis (N by C by gender) with age as time-metric
This document contains the meta-analysis of the surivival models, using age.sex since baseline as the metric of interest and neuroticism by conscientiousness as the predictor of interest.
Code
The following packages were used to generate this table:
library(papaja)
library(tidyverse)
library(metafor)
library(knitr)
library(kableExtra)
library(here)The files needed for this table are available at osf.io/mzfu9 in the Individual Study Output folder.
First we load the individual study analysis objects.
study.names = c("EAS", "HRS", "LBC", "LBLS", "MAP", "MAS", "MIDUS", "OATS", "ROS", "SLS", "WLS")
lapply(here(paste0("mortality/study output/", study.names, "_survival_output.Rdata")), load, .GlobalEnv)meta.data.age.sex <- data.frame()
n <- 0
for(i in study.names){
n <- n+1
x <- get(paste0(i, "_survival_output"))
meta.data.age.sex[n, "study"] <- i
meta.data.age.sex[n, "coef"] <- x$age$model3$coef["z.neur:z.con", "coef"]
meta.data.age.sex[n, "se"] <- x$age$model3$coef["z.neur:z.con", "se(coef)"]
meta.data.age.sex[n, "n"] <- x$age$model3$ntotal
meta.data.age.sex[n, "lowC.male.est"] <- x$HR.N$age.sex$LowC.male$est
meta.data.age.sex[n, "lowC.male.se"] <- x$HR.N$age.sex$LowC.male$se
meta.data.age.sex[n, "hghC.male.est"] <- x$HR.N$age.sex$HghC.male$est
meta.data.age.sex[n, "hghC.male.se"] <- x$HR.N$age.sex$HghC.male$se
meta.data.age.sex[n, "lowC.female.est"] <- x$HR.N$age.sex$LowC.female$est
meta.data.age.sex[n, "lowC.female.se"] <- x$HR.N$age.sex$LowC.female$se
meta.data.age.sex[n, "hghC.female.est"] <- x$HR.N$age.sex$HghC.female$est
meta.data.age.sex[n, "hghC.female.se"] <- x$HR.N$age.sex$HghC.female$se
#calculate hazard ratios at low and high C
lowC.male.HR <- exp(x$HR.N$age.sex$LowC.male$est)
lowC.male.HR.lwr <- exp(x$HR.N$age.sex$LowC.male$est - 1.96*x$HR.N$age.sex$LowC.male$se)
lowC.male.HR.upr <- exp(x$HR.N$age.sex$LowC.male$est + 1.96*x$HR.N$age.sex$LowC.male$se)
meta.data.age.sex[n, "lowC.male.CI"] <- paste0(printnum(lowC.male.HR)," [",printnum(lowC.male.HR.lwr),", ",printnum(lowC.male.HR.upr),"]")
hghC.male.HR <- exp(x$HR.N$age.sex$HghC.male$est)
hghC.male.HR.lwr <- exp(x$HR.N$age.sex$HghC.male$est - 1.96*x$HR.N$age.sex$HghC.male$se)
hghC.male.HR.upr <- exp(x$HR.N$age.sex$HghC.male$est + 1.96*x$HR.N$age.sex$HghC.male$se)
meta.data.age.sex[n, "hghC.male.CI"] <- paste0(printnum(hghC.male.HR),
" [",printnum(hghC.male.HR.lwr),", ",
printnum(hghC.male.HR.upr),"]")
lowC.female.HR <- exp(x$HR.N$age.sex$LowC.female$est)
lowC.female.HR.lwr <- exp(x$HR.N$age.sex$LowC.female$est - 1.96*x$HR.N$age.sex$LowC.female$se)
lowC.female.HR.upr <- exp(x$HR.N$age.sex$LowC.female$est + 1.96*x$HR.N$age.sex$LowC.female$se)
meta.data.age.sex[n, "lowC.female.CI"] <- paste0(printnum(lowC.female.HR),
" [",printnum(lowC.female.HR.lwr),", ",printnum(lowC.female.HR.upr),"]")
hghC.female.HR <- exp(x$HR.N$age.sex$HghC.female$est)
hghC.female.HR.lwr <- exp(x$HR.N$age.sex$HghC.female$est - 1.96*x$HR.N$age.sex$HghC.female$se)
hghC.female.HR.upr <- exp(x$HR.N$age.sex$HghC.female$est + 1.96*x$HR.N$age.sex$HghC.female$se)
meta.data.age.sex[n, "hghC.female.CI"] <- paste0(printnum(hghC.female.HR),
" [",printnum(hghC.female.HR.lwr),", ",printnum(hghC.female.HR.upr),"]")
}
meta.results.age.sex <- rma(yi = coef,
sei = se,
ni = n,
measure = "RR",
slab = study,
data = meta.data.age.sex)#meta-analyze HR at low and high c
meta.lowmale.age.sex <- rma(yi = lowC.male.est,
sei = lowC.male.se,
ni = n,
measure = "RR",
slab = study,
data = meta.data.age.sex)
meta.hghmale.age.sex <- rma(yi = hghC.male.est,
sei = hghC.male.se,
ni = n,
measure = "RR",
slab = study,
data = meta.data.age.sex)
meta.lowfemale.age.sex <- rma(yi = lowC.female.est,
sei = lowC.female.se,
ni = n,
measure = "RR",
slab = study,
data = meta.data.age.sex)
meta.hghfemale.age.sex <- rma(yi = hghC.female.est,
sei = hghC.female.se,
ni = n,
measure = "RR",
slab = study,
data = meta.data.age.sex)
#find plot limits
max.ci.mod1 = max(exp(meta.data.age.sex$coef+1.96*meta.data.age.sex$se))
min.ci.mod1 = min(exp(meta.data.age.sex$coef-1.96*meta.data.age.sex$se))
range.mod1 = max.ci.mod1-min.ci.mod1
lower.mod1 = min.ci.mod1-(range.mod1*7)
upper.mod1 = max.ci.mod1+(range.mod1*1.5)
cex.set = .7
#estimate position of extra information
pos.mod1 = min.ci.mod1-lower.mod1
pos.mod1 = pos.mod1/15
pos.mod1 = c(lower.mod1+3*pos.mod1,
lower.mod1+5*pos.mod1,
lower.mod1+7.5*pos.mod1,
lower.mod1+10.5*pos.mod1,
lower.mod1+13*pos.mod1)
#extract weighted values at -1 and +1 sd
meta_est_lowc_male_mod1 = printnum(exp(meta.lowmale.age.sex$b))
meta_lwr_lowc_male_mod1 = printnum(exp(meta.lowmale.age.sex$ci.lb))
meta_upr_lowc_male_mod1 = printnum(exp(meta.lowmale.age.sex$ci.ub))
meta_lowc_male_ci_mod1 = paste0(meta_est_lowc_male_mod1, " [",
meta_lwr_lowc_male_mod1, ", ",
meta_upr_lowc_male_mod1, "]")
meta_est_hghc_male_mod1 = printnum(exp(meta.hghmale.age.sex$b))
meta_lwr_hghc_male_mod1 = printnum(exp(meta.hghmale.age.sex$ci.lb))
meta_upr_hghc_male_mod1 = printnum(exp(meta.hghmale.age.sex$ci.ub))
meta_hghc_male_ci_mod1 = paste0(meta_est_hghc_male_mod1, " [",
meta_lwr_hghc_male_mod1, ", ",
meta_upr_hghc_male_mod1, "]")
meta_est_lowc_female_mod1 = printnum(exp(meta.lowfemale.age.sex$b))
meta_lwr_lowc_female_mod1 = printnum(exp(meta.lowfemale.age.sex$ci.lb))
meta_upr_lowc_female_mod1 = printnum(exp(meta.lowfemale.age.sex$ci.ub))
meta_lowc_female_ci_mod1 = paste0(meta_est_lowc_female_mod1, " [",
meta_lwr_lowc_female_mod1, ", ",
meta_upr_lowc_female_mod1, "]")
meta_est_hghc_female_mod1 = printnum(exp(meta.hghfemale.age.sex$b))
meta_lwr_hghc_female_mod1 = printnum(exp(meta.hghfemale.age.sex$ci.lb))
meta_upr_hghc_female_mod1 = printnum(exp(meta.hghfemale.age.sex$ci.ub))
meta_hghc_female_ci_mod1 = paste0(meta_est_hghc_female_mod1, " [",
meta_lwr_hghc_female_mod1, ", ",
meta_upr_hghc_female_mod1, "]")#forest plot
meta.data.age.sex$study = gsub("LBC", "LBC1936", meta.data.age.sex$study)
forest(meta.results.age.sex,
xlim = c(lower.mod1, upper.mod1),
cex = cex.set,
slab = meta.data.age.sex$study,
transf=exp,
refline = 1,
ilab = meta.data.age.sex[,c("n",
"lowC.male.CI",
"hghC.male.CI",
"lowC.female.CI",
"hghC.female.CI")],
ilab.xpos = pos.mod1)
# #additional text
text(pos.mod1[1], length(study.names)+1.5,
c("N"), cex = cex.set)
text(pos.mod1[2], length(study.names)+1.5,
c("Low Con"), cex = cex.set)
text(pos.mod1[3], length(study.names)+1.5,
c("High Con"), cex = cex.set)
text(pos.mod1[4], length(study.names)+1.5,
c("Low Con"), cex = cex.set)
text(pos.mod1[5], length(study.names)+1.5,
c("High Con"), cex = cex.set)
text((pos.mod1[2]+pos.mod1[3])/2, length(study.names)+2.5,
c("Male"), cex = cex.set)
text((pos.mod1[4]+pos.mod1[5])/2, length(study.names)+2.5,
c("Female"), cex = cex.set)
text((pos.mod1[3]+pos.mod1[4])/2, length(study.names)+3,
"Neuroticism Hazard Ratio", cex = cex.set)
text(lower.mod1, length(study.names)+1.5,
"Study", cex = cex.set, pos = 4)
text(upper.mod1, length(study.names)+1.5,
"Interaction Hazard Ratio [95% CI]", cex = cex.set, pos=2)
text(pos.mod1[2:5], -1,
c(meta_lowc_male_ci_mod1, meta_hghc_male_ci_mod1,
meta_lowc_female_ci_mod1, meta_hghc_female_ci_mod1), cex = cex.set)
text(lower.mod1, -2, pos=4, cex=cex.set,
bquote(paste("(Q = ",
.(formatC(meta.results.age.sex$QE, digits=2, format="f")),
", df = ", .(meta.results.age.sex$k - meta.results.age.sex$p),
", p = ", .(formatC(meta.results.age.sex$QEp, digits=2, format="f")),
"; ", I^2, " = ",
.(formatC(meta.results.age.sex$I2, digits=1, format="f")), "%)")))