Main Effects of Neuroticism
This document organizes and summarizes the main effects of neuroticism on health outcomes, controlling for conscientiousness, across the studies.
Code
The following packages were used to generate this figure:
library(tidyverse)
library(metafor)
library(papaja)
library(here)The files needed for this table are available at osf.io/mzfu9 in the Individual Study Output folder.
First we load the individual study analysis objects.
study.names = c("BASEII", "EAS", "ELSA", "HRS", "ILSE", "LBC",
"LBLS","MAP", "MAS", "MIDUS",
"NAS", "OATS", "ROS", "SLS", "WLS")
load(here("chronic/created data/BASEII_cc_output_nosrh.Rdata"))
load(here("chronic/created data/EAS_cc_nosrh_output.Rdata"))
load(here("chronic/created data/ELSA_cc_output.Rdata"))
load(here("chronic/created data/HRS_cc_output.Rdata"))
load(here("chronic/created data/ILSE_cc_output_nosrh.Rdata"))
load(here("chronic/created data/LBC_cc_output.Rdata"))
load(here("chronic/created data/LBLS_cc_output.Rdata"))
load(here("chronic/created data/MAP_cc_output.Rdata"))
load(here("chronic/created data/MAS_cc_output_nosrh.Rdata"))
load(here("chronic/created data/MIDUS_cc_output_nosrh.Rdata"))
load(here("chronic/created data/nas_cc_output.Rdata"))
load(here("chronic/created data/OATS_cc_output_nosrh.Rdata"))
load(here("chronic/created data/ROS_cc_output.Rdata"))
load(here("chronic/created data/SLS_cc_output.Rdata"))
load(here("chronic/created data/WLS_cc_output.Rdata"))
NAS_cc_output = nas_cc_output
rm(nas_cc_output)We extract the relevant statistics in a loop. (The first author is just learning how to use the purrr package, and so often resorts to loops when under a time constraint.)
We extract the coefficient estimates of neuroticism and the standard errors for each study. We do this separately for the cross-sectional and longitudinal models, so there is one data frame for each.
# cross-sectional models
data_cross_main <- data.frame() # create empty data frame
position <- 0 # start at row 0
for(i in study.names){ # loop through studies
position = position + 1 # increase row by 1
x <- get(paste0(i,"_cc_output")) # get output object
# fill in study-level data
data_cross_main[position, "name"] <- i
#extract coefficient estimates from diabetes model
data_cross_main$neur_est_diab[position] <- x$main_effects$diabetes$cross$coef["z.neur", "Estimate"]
data_cross_main$neur_se_diab[position] <- x$main_effects$diabetes$cross$coef["z.neur", "Std. Error"]
data_cross_main$n_diab[position] <- x$main_effects$diabetes$cross$n
#extract coefficient estimates from high blood pressure model
data_cross_main$neur_est_hbp[position] <- x$main_effects$hbp$cross$coef["z.neur", "Estimate"]
data_cross_main$neur_se_hbp[position] <- x$main_effects$hbp$cross$coef["z.neur", "Std. Error"]
data_cross_main$n_hbp[position] <- x$main_effects$hbp$cross$n
#extract coefficient estimates from heart disease model
if(!is.null(x$main_effects$heart$cross$coef)){
data_cross_main$neur_est_heart[position] <- x$main_effects$heart$cross$coef["z.neur", "Estimate"]
data_cross_main$neur_se_heart[position] <- x$main_effects$heart$cross$coef["z.neur", "Std. Error"]
data_cross_main$n_heart[position] <- x$main_effects$heart$cross$n
}
}
#longitudinal models
data_long_main <- data.frame() # create empty data frame
position <- 0 # start at row 0
for(i in study.names){ # loop through studies
position = position + 1 # increase row by 1
x <- get(paste0(i,"_cc_output")) # get output object
# fill in study-level data
data_long_main[position, "name"] <- i
#extract coefficient estimates from diabetes model
if(!is.null(x$main_effects$diabetes$long$coef)){
data_long_main$neur_est_diab[position] <- x$main_effects$diabetes$long$coef["z.neur", "Estimate"]
data_long_main$neur_se_diab[position] <- x$main_effects$diabetes$long$coef["z.neur", "Std. Error"]
data_long_main$n_diab[position] <- x$main_effects$diabetes$long$n
}
#extract coefficient estimates from high blood pressure model
data_long_main$neur_est_hbp[position] <- x$main_effects$hbp$long$coef["z.neur", "Estimate"]
data_long_main$neur_se_hbp[position] <- x$main_effects$hbp$long$coef["z.neur", "Std. Error"]
data_long_main$con_est_hbp[position] <- x$main_effects$hbp$long$coef["z.con", "Estimate"]
data_long_main$con_se_hbp[position] <- x$main_effects$hbp$long$coef["z.con", "Std. Error"]
data_long_main$n_hbp[position] <- x$main_effects$hbp$long$n
#extract coefficient estimates from heart disease model
if(!is.null(x$main_effects$heart$long$coef)){
data_long_main$neur_est_heart[position] <- x$main_effects$heart$long$coef["z.neur", "Estimate"]
data_long_main$neur_se_heart[position] <- x$main_effects$heart$long$coef["z.neur", "Std. Error"]
data_long_main$n_heart[position] <- x$main_effects$heart$long$n
}
}Next, we use the tools of meta-analysis to estimate the weighted average effect and heterogeneity for each outcome.
# high blood pressure - neuroticism
meta.cross.hbp.neur <- metafor::rma(yi = neur_est_hbp,
sei = neur_se_hbp,
ni = data_cross_main$n_diab,
measure = "OR",
slab = name,
method="REML",
data = data_cross_main)
# diabetes - neuroticism
meta.cross.diab.neur <- metafor::rma(yi = neur_est_diab,
sei = neur_se_diab,
ni = data_cross_main$n_diab,
measure = "OR",
slab = name,
method="REML",
data = data_cross_main)
# heart disease - neuroticism
meta.cross.heart.neur <- metafor::rma(yi = neur_est_heart,
sei = neur_se_heart,
ni = data_cross_main$n_diab,
measure = "OR",
slab = name,
method="REML",
data = data_cross_main)
# ------ meta-analysis: longitudinal ------
# high blood pressure - neuroticism
meta.long.hbp.neur <- metafor::rma(yi = neur_est_hbp,
sei = neur_se_hbp,
ni = data_long_main$n_diab,
measure = "OR",
slab = name,
method="REML",
data = data_long_main)
# diabetes - neuroticism
meta.long.diab.neur <- metafor::rma(yi = neur_est_diab,
sei = neur_se_diab,
ni = data_long_main$n_diab,
measure = "OR",
slab = name,
method="REML",
data = data_long_main)
# heart disease - neuroticism
meta.long.heart.neur <- metafor::rma(yi = neur_est_heart,
sei = neur_se_heart,
ni = data_long_main$n_diab,
measure = "OR",
slab = name,
method="REML",
data = data_long_main)Finally, we put the information into a single forest plot.
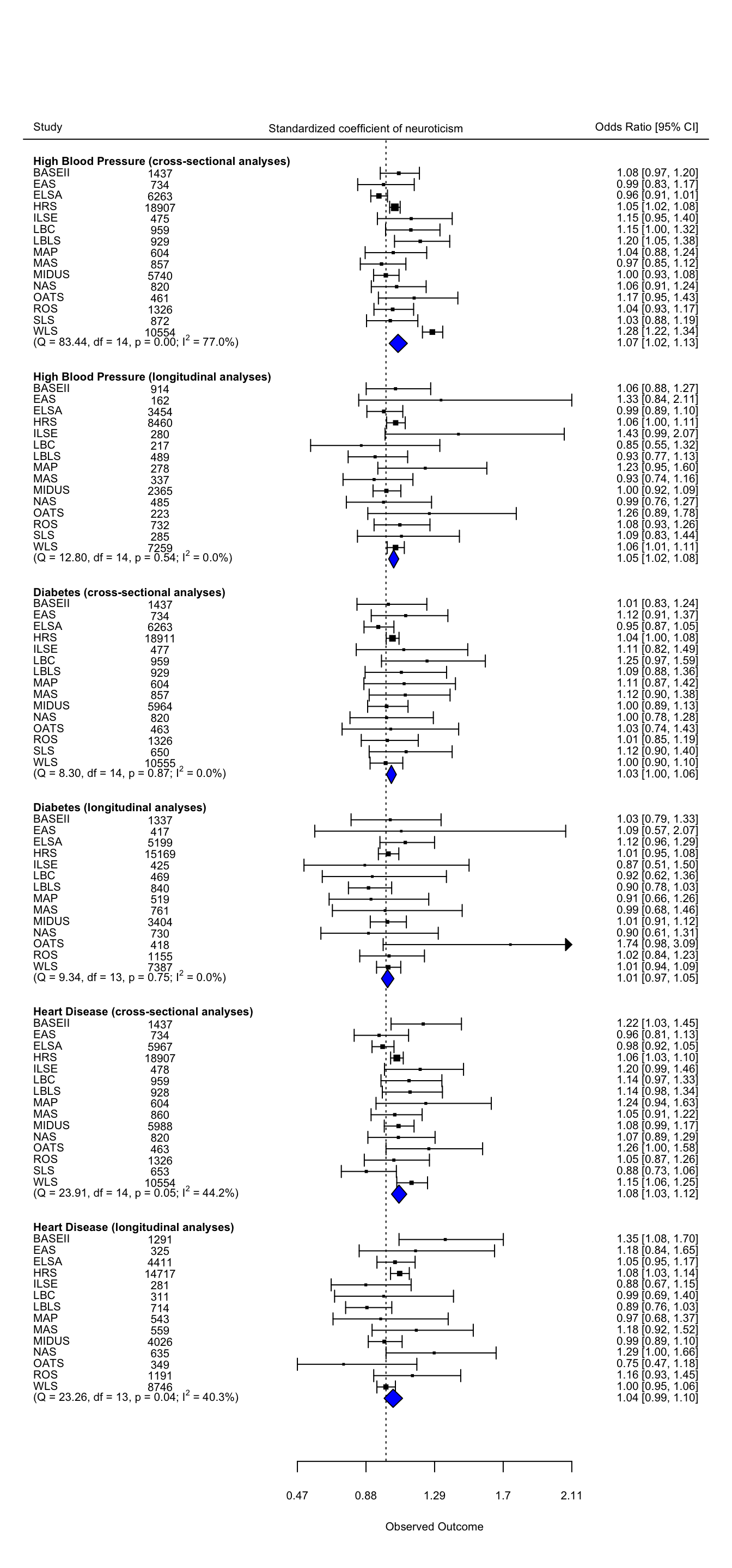
neur.data = data.frame(study = c(rep(data_cross_main$name, 3),rep(data_long_main$name, 3)),
outcome = c(rep("diabetes", nrow(data_cross_main)),
rep("high blood pressure", nrow(data_cross_main)),
rep("heart disease", nrow(data_cross_main)),
rep("diabetes", nrow(data_long_main)),
rep("high blood pressure", nrow(data_long_main)),
rep("heart disease", nrow(data_long_main))),
model = c(rep("cross", nrow(data_cross_main)*3),
rep("long", nrow(data_cross_main)*3)),
estimate = c(data_cross_main$neur_est_diab,
data_cross_main$neur_est_hbp,
data_cross_main$neur_est_heart,
data_long_main$neur_est_diab,
data_long_main$neur_est_hbp,
data_long_main$neur_est_heart),
se = c(data_cross_main$neur_se_diab,
data_cross_main$neur_se_hbp,
data_cross_main$neur_se_heart,
data_long_main$neur_se_diab,
data_long_main$neur_se_hbp,
data_long_main$neur_se_heart),
position = c(data_cross_main$n_diab,
data_cross_main$n_hbp,
data_cross_main$n_heart,
data_long_main$n_diab,
data_long_main$n_hbp,
data_long_main$n_heart))
neur.data = neur.data %>%
filter(!is.na(estimate)) %>%
mutate(outcome = factor(outcome,
levels = c("high blood pressure", "diabetes", "heart disease"))) %>%
arrange(outcome, model, study)
#find plot limits
max.ci.mod1 = neur.data %>%
mutate(ci = exp(estimate+1.96*se)) %>%
arrange(desc(ci))
max.ci.mod1 = max.ci.mod1[2, "ci"]
#max.ci.mod1 = max(exp(neur.data$estimate+1.96*neur.data$se))
min.ci.mod1 = min(exp(neur.data$estimate-1.96*neur.data$se))
range.mod1 = max.ci.mod1-min.ci.mod1
lower.mod1 = min.ci.mod1-(range.mod1)
upper.mod1 = max.ci.mod1+(range.mod1)*.5
cex.value = .65
#estimate position of extra information
pos.mod1 = min.ci.mod1-lower.mod1
pos.mod1 = pos.mod1/2
pos.mod1 = c(lower.mod1+1*pos.mod1)
# rows
rows.heart.long = which(neur.data$outcome == "heart disease" & neur.data$model == "long")
rows.heart.long = c(1, length(rows.heart.long))
rows.heart.cross = which(neur.data$outcome == "heart disease" & neur.data$model == "cross")
rows.heart.cross = c(rows.heart.long[2] + 5, rows.heart.long[2] + 4 + length(rows.heart.cross))
rows.diabetes.long = which(neur.data$outcome == "diabetes" & neur.data$model == "long")
rows.diabetes.long = c(rows.heart.cross[2] + 5, rows.heart.cross[2] + 4 + length(rows.diabetes.long))
rows.diabetes.cross = which(neur.data$outcome == "diabetes" & neur.data$model == "cross")
rows.diabetes.cross = c(rows.diabetes.long[2] + 5, rows.diabetes.long[2] + 4 + length(rows.diabetes.cross))
rows.hbp.long = which(neur.data$outcome == "high blood pressure" & neur.data$model == "long")
rows.hbp.long = c(rows.diabetes.cross[2] + 5, rows.diabetes.cross[2] + 4 + length(rows.hbp.long))
rows.hbp.cross = which(neur.data$outcome == "high blood pressure" & neur.data$model == "cross")
rows.hbp.cross = c(rows.hbp.long[2] + 5, rows.hbp.long[2] + 4 + length(rows.hbp.cross))
#forest plot
forest(neur.data$estimate, neur.data$se^2,
xlim = c(lower.mod1, upper.mod1),
alim = c(min.ci.mod1, max.ci.mod1),
cex = cex.value, slab = neur.data$study,
transf=exp,
refline = 1,
ylim = c(-1, max(rows.hbp.cross)+5),
rows = c(rows.hbp.cross[2]:rows.hbp.cross[1],
rows.hbp.long[2]:rows.hbp.long[1],
rows.diabetes.cross[2]:rows.diabetes.cross[1],
rows.diabetes.long[2]:rows.diabetes.long[1],
rows.heart.cross[2]:rows.heart.cross[1],
rows.heart.long[2]:rows.heart.long[1]),
ilab = cbind(neur.data$position),
ilab.xpos = pos.mod1)
### add summary polygons
addpoly(meta.long.diab.neur, row=rows.diabetes.long[1]-1,
cex = cex.value, transf =exp, mlab="", col = "blue")
addpoly(meta.cross.diab.neur, row=rows.diabetes.cross[1]-1,
cex = cex.value, transf =exp, mlab="", col = "blue")
addpoly(meta.long.hbp.neur, row=rows.hbp.long[1]-1,
cex = cex.value, transf =exp, mlab="", col = "blue")
addpoly(meta.cross.hbp.neur, row=rows.hbp.cross[1]-1,
cex = cex.value, transf =exp, mlab="", col = "blue")
addpoly(meta.long.heart.neur, row=rows.heart.long[1]-1,
cex = cex.value, transf =exp, mlab="", col = "blue")
addpoly(meta.cross.heart.neur, row=rows.heart.cross[1]-1,
cex = cex.value, transf =exp, mlab="", col = "blue")
### add text with Q-value, dfs, p-value, and I^2 statistic for subgroups
text(lower.mod1, rows.diabetes.long[1]-1, pos=4, cex = cex.value,
bquote(paste("(Q = ",
.(formatC(meta.long.diab.neur$QE, digits=2, format="f")),
", df = ", .(meta.long.diab.neur$k - meta.long.diab.neur$p),
", p = ", .(formatC(meta.long.diab.neur$QEp, digits=2, format="f")), "; ", I^2, " = ",
.(formatC(meta.long.diab.neur$I2, digits=1, format="f")), "%)")))
text(lower.mod1, rows.diabetes.cross[1]-1, pos=4, cex = cex.value,
bquote(paste("(Q = ",
.(formatC(meta.cross.diab.neur$QE, digits=2, format="f")),
", df = ", .(meta.cross.diab.neur$k - meta.cross.diab.neur$p),
", p = ", .(formatC(meta.cross.diab.neur$QEp, digits=2, format="f")), "; ", I^2, " = ",
.(formatC(meta.cross.diab.neur$I2, digits=1, format="f")), "%)")))
text(lower.mod1, rows.hbp.long[1]-1, pos=4, cex = cex.value,
bquote(paste("(Q = ",
.(formatC(meta.long.hbp.neur$QE, digits=2, format="f")),
", df = ", .(meta.long.hbp.neur$k - meta.long.hbp.neur$p),
", p = ", .(formatC(meta.long.hbp.neur$QEp, digits=2, format="f")), "; ", I^2, " = ",
.(formatC(meta.long.hbp.neur$I2, digits=1, format="f")), "%)")))
text(lower.mod1, rows.hbp.cross[1]-1, pos=4, cex = cex.value,
bquote(paste("(Q = ",
.(formatC(meta.cross.hbp.neur$QE, digits=2, format="f")),
", df = ", .(meta.cross.hbp.neur$k - meta.cross.hbp.neur$p),
", p = ", .(formatC(meta.cross.hbp.neur$QEp, digits=2, format="f")), "; ", I^2, " = ",
.(formatC(meta.cross.hbp.neur$I2, digits=1, format="f")), "%)")))
text(lower.mod1, rows.heart.long[1]-1, pos=4, cex = cex.value,
bquote(paste("(Q = ",
.(formatC(meta.long.heart.neur$QE, digits=2, format="f")),
", df = ", .(meta.long.heart.neur$k - meta.long.heart.neur$p),
", p = ", .(formatC(meta.long.heart.neur$QEp, digits=2, format="f")), "; ", I^2, " = ",
.(formatC(meta.long.heart.neur$I2, digits=1, format="f")), "%)")))
text(lower.mod1, rows.heart.cross[1]-1, pos=4, cex = cex.value,
bquote(paste("(Q = ",
.(formatC(meta.cross.heart.neur$QE, digits=2, format="f")),
", df = ", .(meta.cross.heart.neur$k - meta.cross.heart.neur$p),
", p = ", .(formatC(meta.cross.heart.neur$QEp, digits=2, format="f")), "; ", I^2, " = ",
.(formatC(meta.cross.heart.neur$I2, digits=1, format="f")), "%)")))
###additional text
text(lower.mod1, rows.hbp.cross[2] + 4,
"Study", cex = cex.value, pos = 4)
text(upper.mod1, rows.hbp.cross[2] + 4,
"Odds Ratio [95% CI]", cex = cex.value, pos=2)
text((lower.mod1+upper.mod1)/2, rows.hbp.cross[2] + 4,
"Standardized coefficient of neuroticism", cex = cex.value)
text(lower.mod1, rows.diabetes.cross[2] + 1,
"Diabetes (cross-sectional analyses)", cex = cex.value, pos = 4, font = 2)
text(lower.mod1, rows.diabetes.long[2] + 1,
"Diabetes (longitudinal analyses)", cex = cex.value, pos = 4, font = 2)
text(lower.mod1, rows.hbp.cross[2] + 1,
"High Blood Pressure (cross-sectional analyses)", cex = cex.value, pos = 4, font = 2)
text(lower.mod1, rows.hbp.long[2] + 1,
"High Blood Pressure (longitudinal analyses)", cex = cex.value, pos = 4, font = 2)
text(lower.mod1, rows.heart.cross[2] + 1,
"Heart Disease (cross-sectional analyses)", cex = cex.value, pos = 4, font = 2)
text(lower.mod1, rows.heart.long[2] + 1,
"Heart Disease (longitudinal analyses)", cex = cex.value, pos = 4, font = 2)