Power analysis for heterogeneity in meta-analysis
This document contains the power analysis for the primary models of interest. For each model, we display the power to detect significant \(\tau\) values, or different standard deviations of between-study effects (reported in the metric of odds ratios). We also mark the level of \(tau\) estimated by our model and the power to detect this effect. We note here that we are well powered to detect between-study variability that is commonly found in psychology (van Erp, Verhagen, Grasman, & Wagenmakers, 2017).
Many thanks to Brenton Wiernik, who developed the code to calculate power based on our analyses!
The following packages were used to generate this figure:
Cross-sectional
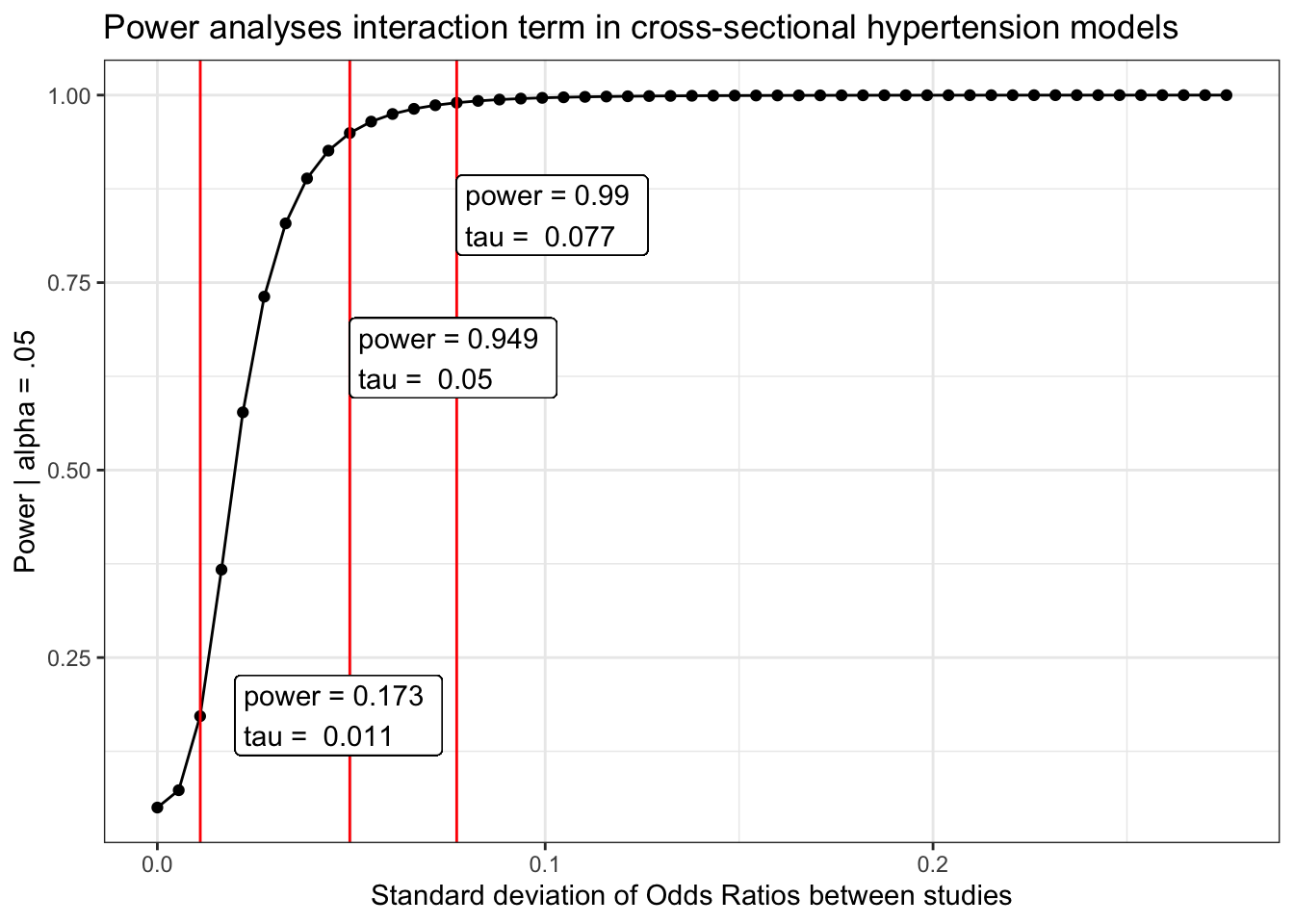
Hypertension/high blood pressure
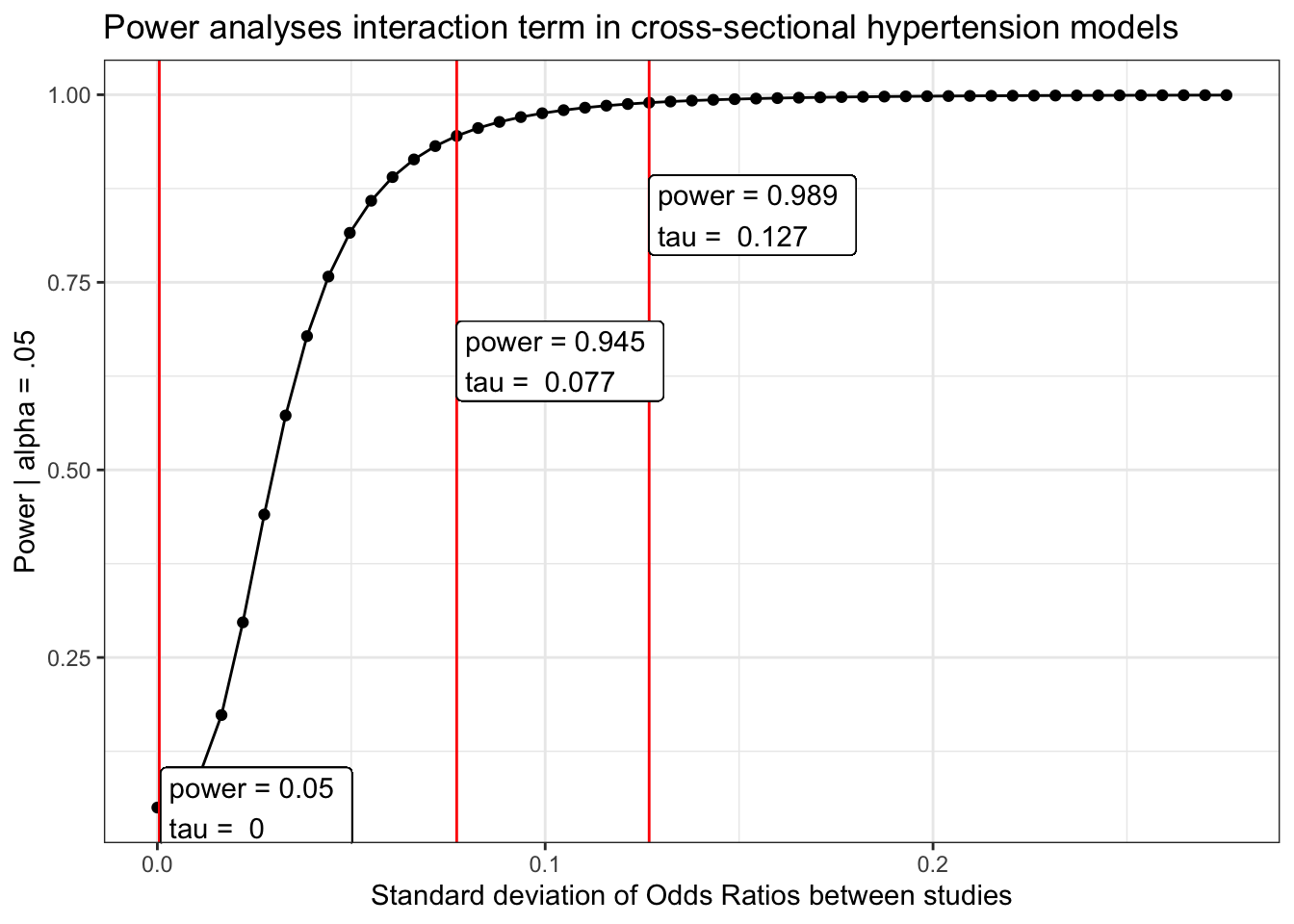
Diabetes
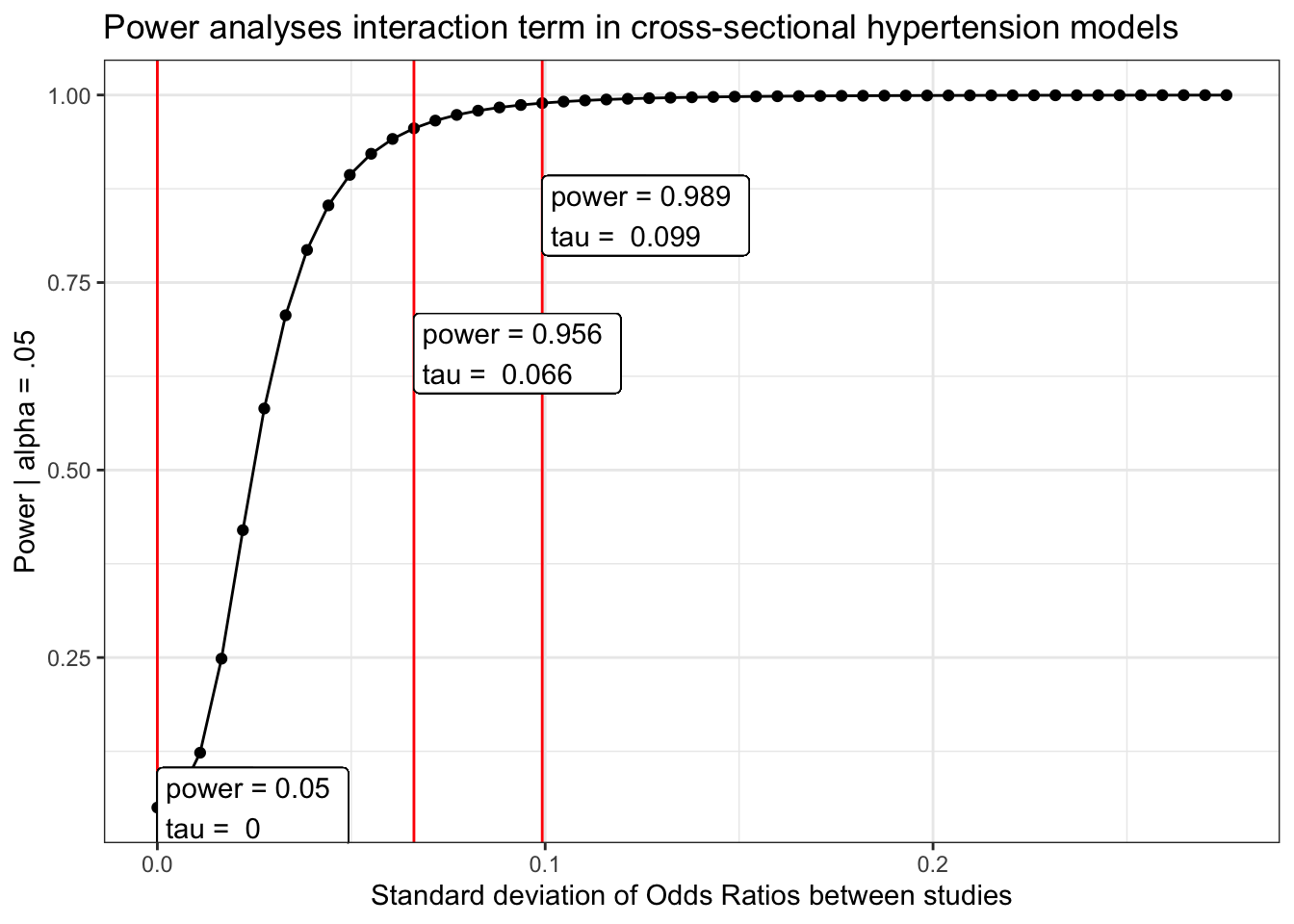
Heart disease
Longitudinal
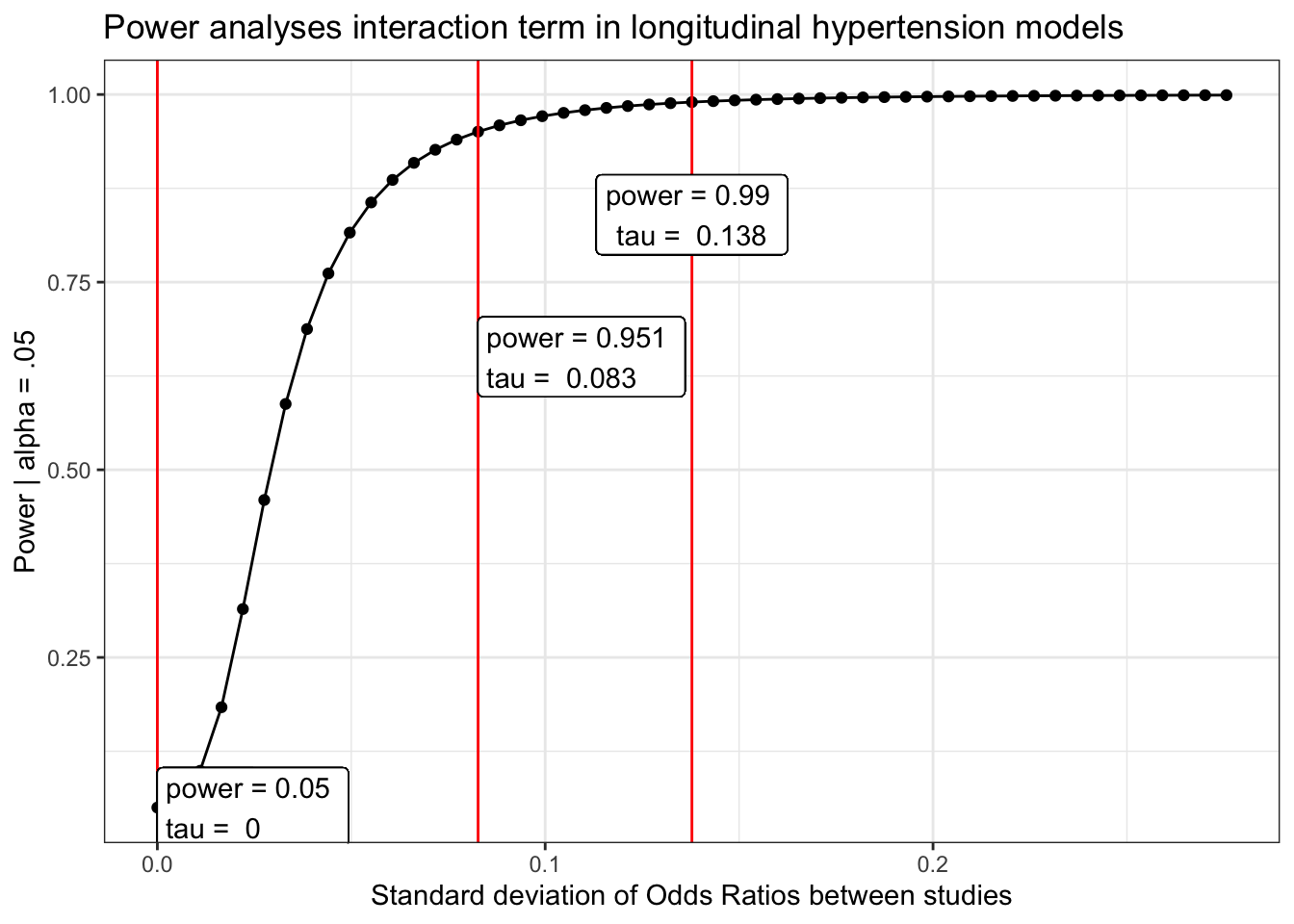
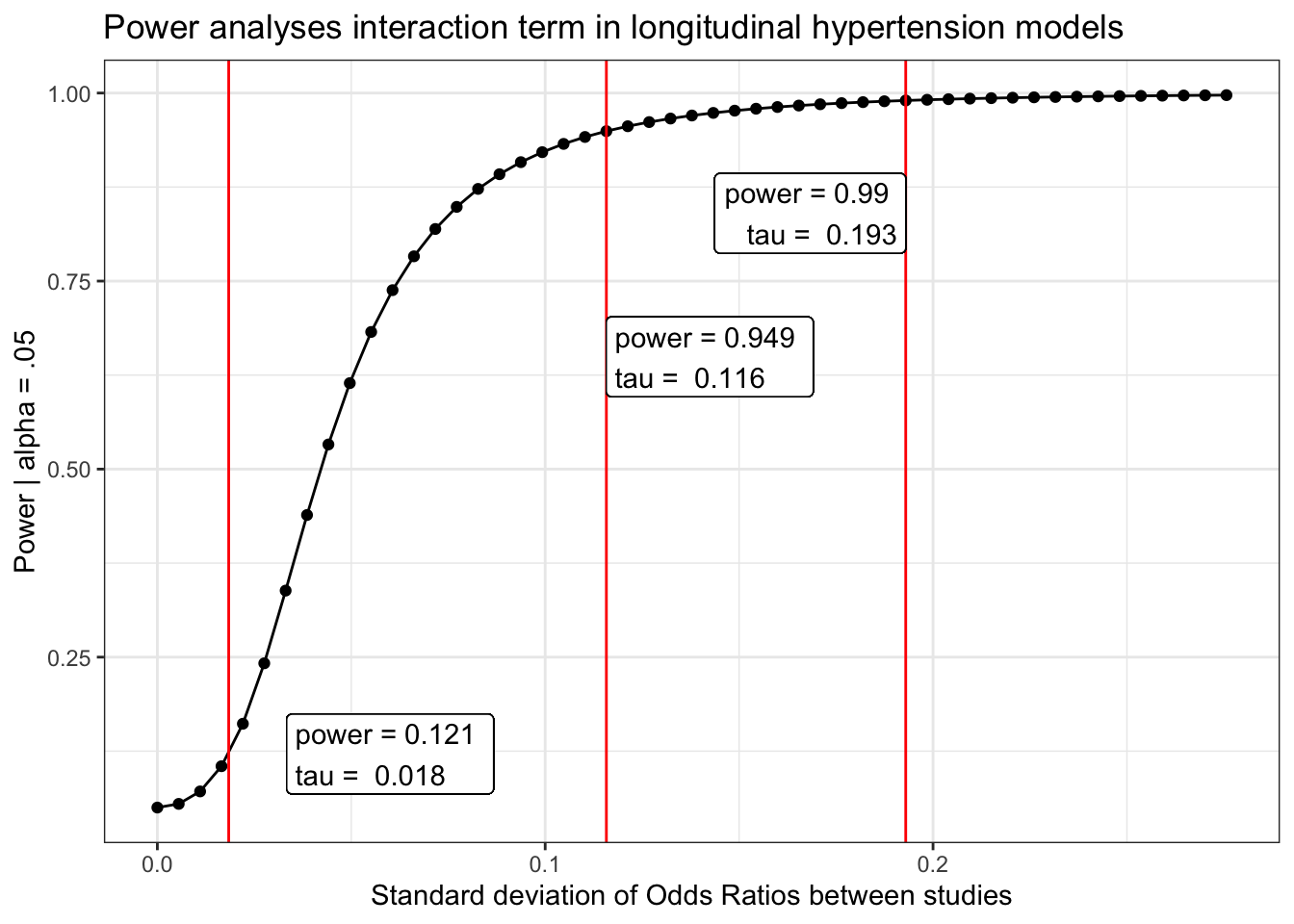
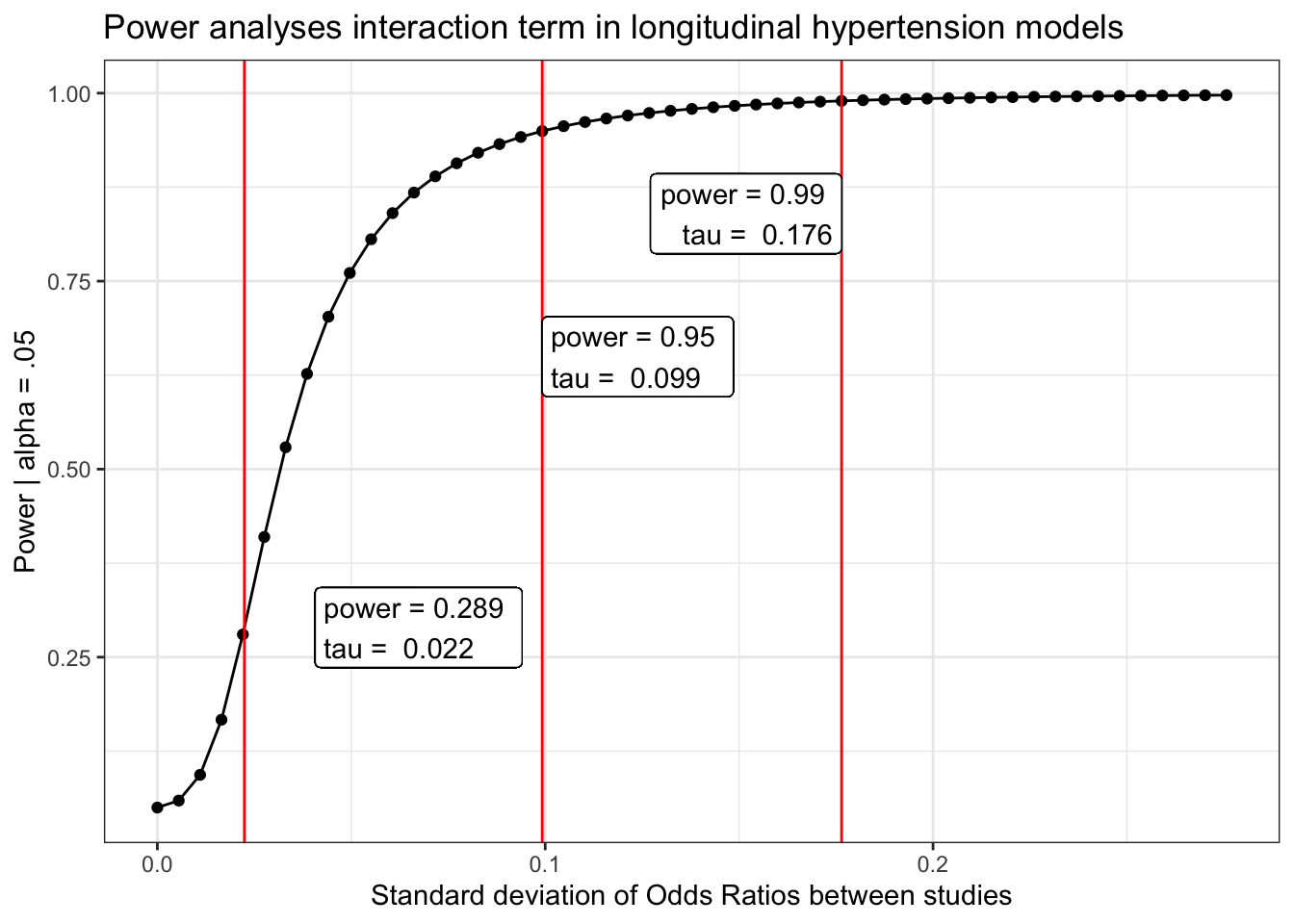
Hypertension/high blood pressure
Diabetes
Heart disease
Code
# Based on methods described by Hedges and Pigott, https://doi.org/10.1037/1082-989X.9.4.426, p. 438
# Written 2019-10-25 by Brenton M. Wiernik
# Licensed GPL v3.0
power_curve <- function(object, tau, level = .95) {
if (! inherits(object, "rma.uni")) stop("'object' must be an object of class 'rma.uni'")
tau2 <- tau^2
obs_tau2 <- object$tau2
obs_tau <- sqrt(obs_tau2)
obs_H2 <- object$H2
s2 <- obs_tau2 / (obs_H2 - 1)
df <- object$k - object$p
crit <- qchisq(.95, df)
wi <- 1 / object$vi
a <- sum(wi) - sum(wi^2) / sum(wi)
mu_q <- function(tau2) a * tau2 + df
b <- function(tau2) df + 2 * a * tau2 +
(sum(wi^2) - 2 * sum(wi^3) / sum(wi) +
sum(wi^2)^2 / sum(wi)^2) * tau2^2
var_q <- function(tau2) 2 * b(tau2)
r <- function(tau2) var_q(tau2) / (2 * mu_q(tau2))
s <- function(tau2) 2 * mu_q(tau2)^2 / var_q(tau2)
H <- function(tau2) pchisq(crit / r(tau2), s(tau2))
power <- 1 - H(tau2)
out <- list(obs_tau = obs_tau, power = data.frame(tau = tau, power = power))
return(out)
}The following packages were used to generate this figure:
library(tidyverse)
library(metafor)
library(papaja)
library(here)tau = seq(0, .5, .01) # possible values of the standard deviation of effect sizesCross-sectional
Hypertension/high blood pressure
load(here("chronic/meta output/hbp_cross.Rdata"))
power_hbp_cross = power_curve(meta.int.mod3, tau, level = .95)
obs = power_curve(meta.int.mod3, power_hbp_cross$obs_tau, level =.95)$power[1,2]
#for which tau are we powered?
pwr.95=.95
pwr.99=.99
tau.95 = which(abs(power_hbp_cross$power$power-pwr.95)==min(abs(power_hbp_cross$power$power-pwr.95)))
pwr.95 = power_hbp_cross$power$power[tau.95]
tau.95 = power_hbp_cross$power$tau[tau.95]
tau.99 = which(abs(power_hbp_cross$power$power-pwr.99)==min(abs(power_hbp_cross$power$power-pwr.99)))
pwr.99 = power_hbp_cross$power$power[tau.99]
tau.99 = power_hbp_cross$power$tau[tau.99]
power_hbp_cross$power %>%
mutate(tau_d = tau/(pi/sqrt(3))) %>%
ggplot(aes(x = tau_d, y = power)) +
geom_point()+
geom_line()+
geom_vline(aes(xintercept = power_hbp_cross$obs_tau/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.95/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.99/(pi/sqrt(3))), color = "red") +
geom_label(aes(x = power_hbp_cross$obs_tau, y = obs,
label = paste("power =", round(obs, 3),
"\ntau = ", round( power_hbp_cross$obs_tau/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.95/(pi/sqrt(3)), y = pwr.95-.3,
label = paste("power =", round(pwr.95, 3),
"\ntau = ", round( tau.95/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.99/(pi/sqrt(3)), y = pwr.99-.15,
label = paste("power =", round(pwr.99, 3),
"\ntau = ", round( tau.99/(pi/sqrt(3)), 3))), hjust = "inward")+
labs(x = "Standard deviation of Odds Ratios between studies",
y = "Power | alpha = .05",
title = "Power analyses interaction term in cross-sectional hypertension models") +
theme_bw()Diabetes
load(here("chronic/meta output/diabetes_cross.Rdata"))
power_diabetes_cross = power_curve(meta.int.mod3, tau, level = .95)
obs = power_curve(meta.int.mod3, power_diabetes_cross$obs_tau, level =.95)$power[1,2]
#for which tau are we powered?
pwr.95=.95
pwr.99=.99
tau.95 = which(abs(power_diabetes_cross$power$power-pwr.95)==min(abs(power_diabetes_cross$power$power-pwr.95)))
pwr.95 = power_diabetes_cross$power$power[tau.95]
tau.95 = power_diabetes_cross$power$tau[tau.95]
tau.99 = which(abs(power_diabetes_cross$power$power-pwr.99)==min(abs(power_diabetes_cross$power$power-pwr.99)))
pwr.99 = power_diabetes_cross$power$power[tau.99]
tau.99 = power_diabetes_cross$power$tau[tau.99]
power_diabetes_cross$power %>%
mutate(tau_d = tau/(pi/sqrt(3))) %>%
ggplot(aes(x = tau_d, y = power)) +
geom_point()+
geom_line()+
geom_vline(aes(xintercept = power_diabetes_cross$obs_tau/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.95/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.99/(pi/sqrt(3))), color = "red") +
geom_label(aes(x = power_diabetes_cross$obs_tau, y = obs,
label = paste("power =", round(obs, 3),
"\ntau = ", round( power_diabetes_cross$obs_tau/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.95/(pi/sqrt(3)), y = pwr.95-.3,
label = paste("power =", round(pwr.95, 3),
"\ntau = ", round( tau.95/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.99/(pi/sqrt(3)), y = pwr.99-.15,
label = paste("power =", round(pwr.99, 3),
"\ntau = ", round( tau.99/(pi/sqrt(3)), 3))), hjust = "inward")+
labs(x = "Standard deviation of Odds Ratios between studies",
y = "Power | alpha = .05",
title = "Power analyses interaction term in cross-sectional hypertension models") +
theme_bw()Heart disease
load(here("chronic/meta output/heart_cross.Rdata"))
power_heart_cross = power_curve(meta.int.mod3, tau, level = .95)
obs = power_curve(meta.int.mod3, power_heart_cross$obs_tau, level =.95)$power[1,2]
#for which tau are we powered?
pwr.95=.95
pwr.99=.99
tau.95 = which(abs(power_heart_cross$power$power-pwr.95)==min(abs(power_heart_cross$power$power-pwr.95)))
pwr.95 = power_heart_cross$power$power[tau.95]
tau.95 = power_heart_cross$power$tau[tau.95]
tau.99 = which(abs(power_heart_cross$power$power-pwr.99)==min(abs(power_heart_cross$power$power-pwr.99)))
pwr.99 = power_heart_cross$power$power[tau.99]
tau.99 = power_heart_cross$power$tau[tau.99]
power_heart_cross$power %>%
mutate(tau_d = tau/(pi/sqrt(3))) %>%
ggplot(aes(x = tau_d, y = power)) +
geom_point()+
geom_line()+
geom_vline(aes(xintercept = power_heart_cross$obs_tau/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.95/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.99/(pi/sqrt(3))), color = "red") +
geom_label(aes(x = power_heart_cross$obs_tau, y = obs,
label = paste("power =", round(obs, 3),
"\ntau = ", round( power_heart_cross$obs_tau/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.95/(pi/sqrt(3)), y = pwr.95-.3,
label = paste("power =", round(pwr.95, 3),
"\ntau = ", round( tau.95/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.99/(pi/sqrt(3)), y = pwr.99-.15,
label = paste("power =", round(pwr.99, 3),
"\ntau = ", round( tau.99/(pi/sqrt(3)), 3))), hjust = "inward")+
labs(x = "Standard deviation of Odds Ratios between studies",
y = "Power | alpha = .05",
title = "Power analyses interaction term in cross-sectional hypertension models") +
theme_bw()Longitudinal
Hypertension/high blood pressure
load(here("chronic/meta output/hbp_long.Rdata"))
power_hbp_long = power_curve(meta.int.mod3, tau, level = .95)
obs = power_curve(meta.int.mod3, power_hbp_long$obs_tau, level =.95)$power[1,2]
#for which tau are we powered?
pwr.95=.95
pwr.99=.99
tau.95 = which(abs(power_hbp_long$power$power-pwr.95)==min(abs(power_hbp_long$power$power-pwr.95)))
pwr.95 = power_hbp_long$power$power[tau.95]
tau.95 = power_hbp_long$power$tau[tau.95]
tau.99 = which(abs(power_hbp_long$power$power-pwr.99)==min(abs(power_hbp_long$power$power-pwr.99)))
pwr.99 = power_hbp_long$power$power[tau.99]
tau.99 = power_hbp_long$power$tau[tau.99]
power_hbp_long$power %>%
mutate(tau_d = tau/(pi/sqrt(3))) %>%
ggplot(aes(x = tau_d, y = power)) +
geom_point()+
geom_line()+
geom_vline(aes(xintercept = power_hbp_long$obs_tau/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.95/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.99/(pi/sqrt(3))), color = "red") +
geom_label(aes(x = power_hbp_long$obs_tau, y = obs,
label = paste("power =", round(obs, 3),
"\ntau = ", round( power_hbp_long$obs_tau/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.95/(pi/sqrt(3)), y = pwr.95-.3,
label = paste("power =", round(pwr.95, 3),
"\ntau = ", round( tau.95/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.99/(pi/sqrt(3)), y = pwr.99-.15,
label = paste("power =", round(pwr.99, 3),
"\ntau = ", round( tau.99/(pi/sqrt(3)), 3))), hjust = "inward")+
labs(x = "Standard deviation of Odds Ratios between studies",
y = "Power | alpha = .05",
title = "Power analyses interaction term in longitudinal hypertension models") +
theme_bw()Diabetes
load(here("chronic/meta output/diabetes_long.Rdata"))
power_diabetes_long = power_curve(meta.int.mod3, tau, level = .95)
obs = power_curve(meta.int.mod3, power_diabetes_long$obs_tau, level =.95)$power[1,2]
#for which tau are we powered?
pwr.95=.95
pwr.99=.99
tau.95 = which(abs(power_diabetes_long$power$power-pwr.95)==min(abs(power_diabetes_long$power$power-pwr.95)))
pwr.95 = power_diabetes_long$power$power[tau.95]
tau.95 = power_diabetes_long$power$tau[tau.95]
tau.99 = which(abs(power_diabetes_long$power$power-pwr.99)==min(abs(power_diabetes_long$power$power-pwr.99)))
pwr.99 = power_diabetes_long$power$power[tau.99]
tau.99 = power_diabetes_long$power$tau[tau.99]
power_diabetes_long$power %>%
mutate(tau_d = tau/(pi/sqrt(3))) %>%
ggplot(aes(x = tau_d, y = power)) +
geom_point()+
geom_line()+
geom_vline(aes(xintercept = power_diabetes_long$obs_tau/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.95/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.99/(pi/sqrt(3))), color = "red") +
geom_label(aes(x = power_diabetes_long$obs_tau, y = obs,
label = paste("power =", round(obs, 3),
"\ntau = ", round( power_diabetes_long$obs_tau/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.95/(pi/sqrt(3)), y = pwr.95-.3,
label = paste("power =", round(pwr.95, 3),
"\ntau = ", round( tau.95/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.99/(pi/sqrt(3)), y = pwr.99-.15,
label = paste("power =", round(pwr.99, 3),
"\ntau = ", round( tau.99/(pi/sqrt(3)), 3))), hjust = "inward")+
labs(x = "Standard deviation of Odds Ratios between studies",
y = "Power | alpha = .05",
title = "Power analyses interaction term in longitudinal hypertension models") +
theme_bw()Heart disease
load(here("chronic/meta output/heart_long.Rdata"))
power_heart_long = power_curve(meta.int.mod3, tau, level = .95)
obs = power_curve(meta.int.mod3, power_heart_long$obs_tau, level =.95)$power[1,2]
#for which tau are we powered?
pwr.95=.95
pwr.99=.99
tau.95 = which(abs(power_heart_long$power$power-pwr.95)==min(abs(power_heart_long$power$power-pwr.95)))
pwr.95 = power_heart_long$power$power[tau.95]
tau.95 = power_heart_long$power$tau[tau.95]
tau.99 = which(abs(power_heart_long$power$power-pwr.99)==min(abs(power_heart_long$power$power-pwr.99)))
pwr.99 = power_heart_long$power$power[tau.99]
tau.99 = power_heart_long$power$tau[tau.99]
power_heart_long$power %>%
mutate(tau_d = tau/(pi/sqrt(3))) %>%
ggplot(aes(x = tau_d, y = power)) +
geom_point()+
geom_line()+
geom_vline(aes(xintercept = power_heart_long$obs_tau/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.95/(pi/sqrt(3))), color = "red") +
geom_vline(aes(xintercept = tau.99/(pi/sqrt(3))), color = "red") +
geom_label(aes(x = power_heart_long$obs_tau, y = obs,
label = paste("power =", round(obs, 3),
"\ntau = ", round( power_heart_long$obs_tau/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.95/(pi/sqrt(3)), y = pwr.95-.3,
label = paste("power =", round(pwr.95, 3),
"\ntau = ", round( tau.95/(pi/sqrt(3)), 3))), hjust = "inward")+
geom_label(aes(x = tau.99/(pi/sqrt(3)), y = pwr.99-.15,
label = paste("power =", round(pwr.99, 3),
"\ntau = ", round( tau.99/(pi/sqrt(3)), 3))), hjust = "inward")+
labs(x = "Standard deviation of Odds Ratios between studies",
y = "Power | alpha = .05",
title = "Power analyses interaction term in longitudinal hypertension models") +
theme_bw()